The threat of bioterrorism is something that a national security laboratory like Sandia is expected to tackle for the nation. Infectious disease diagnosis and treatment is most often associated with the public health domain. Both bioterrorism and naturally occurring infectious disease share enormous potential to threaten human health and economic stability. To protect the nation and world from these dangers, Sandia researchers conduct basic and applied research in infectious disease and pathogenesis, host-pathogen interactions and immunology. The deep understanding that is gained informs the development of a wide array of countermeasures, including detection, medical diagnostics, therapeutics, and vaccines.
Creating knowledge to counter disease
Biology of Pathogens and Host-Pathogen Interactions
Sandia conducts research into how pathogens interact and subvert a host’s immune response to develop the knowledge base needed to create new novel environmental detectors, medical diagnostics systems, therapeutics, and vaccines to counter natural or human-made outbreaks.
Employing Sandia-developed cutting-edge tools to probe and analyze the molecular events involved in host-pathogen interaction, Sandia scientists are advancing state-of-the-art in multiple critical areas:
- Using next generation sequencing to identify genomic pathogenic signatures in complex clinical matrices
- Conducting studies to improve understanding of the biology of pathogens
- Elucidating the interactions of pathogens with the host and the environment
- Developing methods of analyzing and characterizing pathogens in clinical samples and tissues
- Creating models to capture multi-omic data (or data from genomics, proteomics, and metabolomics) to build interaction networks
Countermeasure Deployment
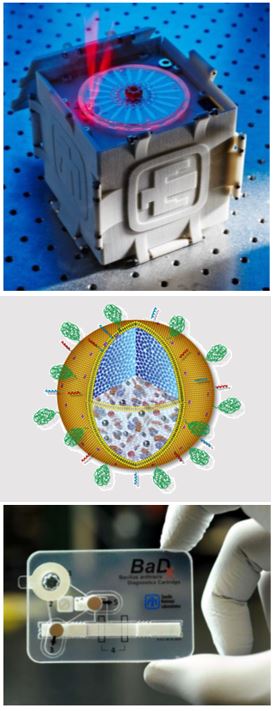
Applying new knowledge of infectious disease, pathogenesis and pathogen-host interactions, Sandia researchers are developing a range of countermeasures. These include fully integrated devices for detecting bioagents in clinical and environmental samples and automated platforms for sample preparation.
Additional work has focused on biological countermeasures such as engineered nanoparticles that can serve as nanodelivery platforms for vaccines and antibiotics.
Many of our patented (and patent-pending) innovations have been licensed to companies for development of commercial devices and vaccines.
Examples of Recent Research
Host-Response to Infectious Disease
Understanding the response of the host to infectious disease is a key piece of the scientific puzzle and could allow us to detect diseases before symptoms appear and has the potential to provide more accurate staging of the disease.
Host-directed therapies enable an alternative route to protecting against infection. These approaches can include:
- Manipulating host immune factors exploited by the pathogen for infection
- Amplifying or prolonging an appropriate host immune response
- Eliciting an unusual host response that is effective.
Moreover, a thorough understanding of the host response could lead to new therapeutic options, particularly when a disease agent is resistant to traditional antimicrobial therapeutics.
Researchers at Sandia, in collaboration with researchers at Lawrence Livermore National Laboratories, have been exploring the host response to one such resistant group of pathogens. Burkholderia spp. B. pseudomallei and B. mallei are closely related Gram-negative bacteria that cause highly lethal disease in humans and animals. Antibiotic treatment is often unsuccessful due to the intrinsic resistance of both pathogens to a wide variety of antibiotics. Both species can persist and replicate within host cells, enabling them to evade many host defense mechanisms.
Recent studies focus on the initial interactions between bacterial pathogens of biodefense concern and the human host, with the idea that improved understanding of these interactions will inform development of useful countermeasures. Researchers are particularly interested in approaches that enable analysis of both host and pathogen in the context of an active infection.
Taking advantage of the recent revolution in Next Generation Sequencing (NGS) technology and its application to transcriptome profiling (quantitative analysis of the transcriptional output of all genes in a cell, tissue, or organism, known as RNA-Seq), we have developed sample preparation methods that enable efficient RNA-Seq analysis of both host and pathogen (dual RNA-Seq) during infection. Through application of these methods, we have carried out dual RNA-Seq analysis of in vitro infection models in which human primary cells (macrophages or airway epithelial cells) are exposed to B. pseudomallei or an attenuated surrogate pathogen, B. thailandensis.
Sandia researchers have discovered the roles macrophages and airway epithelial cells play in induction of pathways mediating immune response, cell-cell and cell-matrix interactions as well as biosynthesis of known virulence factors. The team is currently attempting to identify intervention points at which the host-pathogen interactions within the infection models may be manipulated in order to favor host defense.
Real-Time Autonomous Surveillance for Vector-Borne Pathogens

Mosquito-borne pathogens are a growing menace worldwide. Malaria and dengue significantly burden both civilian and military populations in the tropics, and emerging arboviruses pose health and security risks across the continental U.S. (e.g., West Nile virus, chikungunya virus, and now Zika virus). While the risks posed by arboviruses are well known, surveillance for vector-borne diseases is constrained by limited budgets, and a reliance on labor-intensive techniques for sample collection and analysis.
The Smart Trap was developed as an automated surveillance device that addresses these problems in collaboration with University of California-Davis researchers.
Funded by U.S. Defense Threat Reduction Agency’s biosurveillance ecosystem (BSVE) in collaboration with UC Davis, we have developed a prototype device that automates a novel approach to detecting arboviruses in a field setting, based upon the sugar feeding behavior of mosquitoes. The traps are intended to be inexpensive enough to deploy in, allowing daily reports at multiple locations within a region of interest.

- Trap can detect as low as 1 plaque forming unit (PFU) of a six- virus panel from sugar feeding mosquitos with low false-positive rate in 30 min.
- Trap is designed for mass manufacture at low cost.
- Trap can be reconfigured to target other pathogens.
- Networked biosurveillance using a mesh network
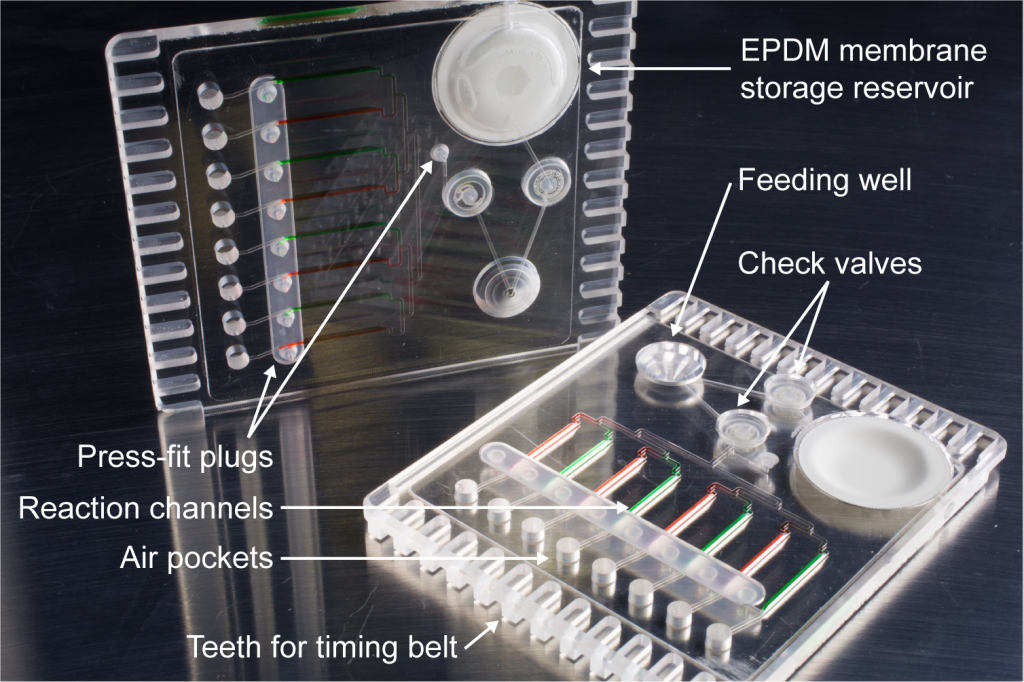
The system uses molecular assays implemented in a microfluidic chip. Mosquitoes feed upon plant-based sugar on a daily basis for energy. Infected mosquitoes can deposit anywhere from 1-10,000 PFUs of virus on a sugar bait, which we detect by a novel reverse transcription loop-mediated isothermal amplification (RT-LAMP) detection assay with fluorescence detection.
The traditional RT-LAMP assays are enhanced by a novel method called “QUASR” (Quenching of Unincorporated Amplification Signal Reporters), originally demonstrated for arbovirus detection. QUASR achieves bright, unambiguous fluorescence signals that can be interpreted with simple instrumentation including a smart phone CMOS camera, and offers target-specific, multiplexable signals which greatly reduce the detection of false-positive amplification. QUASR improves detection in complex matrices such as 10% whole blood, compared to traditional LAMP detection techniques such as turbidity or intercalating dyes. QUASR also performs better than intercalating dyes in plastic microfluidic channels with high signal-to-background ratio.
Antibiotic Resistance
Antibiotic resistance and multidrug resistance is a growing problem around the globe. Each year in the U.S. alone, at least 23,000 people die from infections caused by antibiotic-resistant bacteria, according to the Centers for Disease Control and Prevention.

In order to address this challenge, Sandia has active research efforts in both developing fundamental understanding of how microbes evolve and acquire resistance moieties. Carbapenems are a class of antimicrobial that has been a powerful tool in the fight against multidrug resistant pathogens. That power may be dwindling in light of the emergence of carbapenem resistant strains of Klebsiella pneumoniae.
Sandia has also been exploring additional methods to circumvent antibiotic resistance in pathogens. One way bacteria develop resistance to many different antibiotics is by producing pumps that spit out antibiotics before they can do damage. In one strategy, researchers are teasing out the details of how one antibiotic pump works. The eventual goal is to develop new drugs to plug the pump, or to break it, so it cannot spit out antibiotics, perhaps restoring their effectiveness.
K. pneumoniae is a gram-negative bacterium that is resistant to carbapenem and it typically encountered as a hospital-acquired infection. K. pneumoniae can accumulate antibiotic resistance genes through transfer of genetic elements from other strains.

Using next-generation sequencing and advanced bioinformatics tools, the Sandia team has assembled and presented the complete genome for the first time. The team studied the genome and, using bioinformatics tools designed to identify genomic islands, were able to identify and characterize these mobile genetic elements. Using this and phylogenetic methods, researchers were able to shed light on the evolution of an extensive repertoire of resistance genes, including some 8 carbapenem-active moieties and 15 additional antibiotic resistance enzymes. Explore our bioinformatics site to explore the full genome.
A breakthrough in the antibiotic pump research was the development of new computational tools to come up with an improved structure that could be used in molecular simulations to understand the mechanism of the pump.
Using computer modeling, researchers from Sandia and the University of Illinois at Urbana-Champaign are helping to develop the means to prevent some of the deaths resulting from antibiotic resistant pathogens. Funded in part through Sandia’s Campus Executive Program, the goal is to develop new drugs to plug the pump preventing the bacterium from protecting itself from the antibiotics.
The specific pump researchers studied, called EmrE, comes from E. coli, common bacteria that occasionally cause food poisoning. The pump recognizes and removes moderately oily, positively charged small molecules, which describes many common antibiotics, including streptomycin, doxycycline and chloramphenicol.
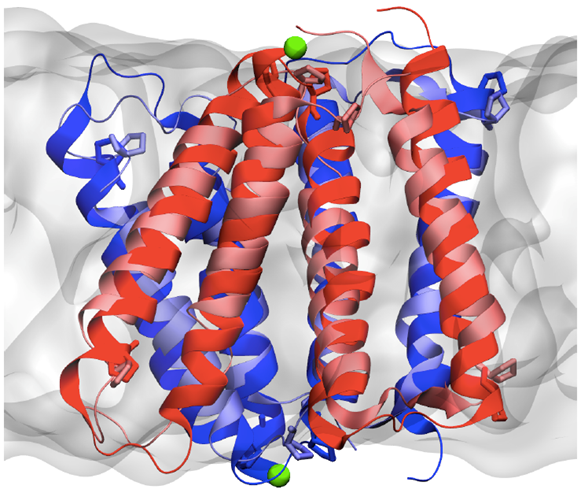
Using combined experimental data from a variety of common biophysical methods such as X-ray crystallography, cryo-electron microscopy and electron paramagnetic resonance spectroscopy, as well as deep knowledge of amino acids, the building blocks of proteins, the team was able to produce a high-resolution structure of the pump.
The team ran simulations to seek out potential drugs that could block the pump and identified critical amino acids that serve as a lock to ensure that the pump does not let go of the protons. The knowledge about the “lock” could lead to new antibiotics that are not pumped out of the bacteria and can perform their job of destroying the infection.