Anne Ruffing
Principal Member of the Technical Staff

Principal Member of the Technical Staff
(505) 850-4999
Sandia National Laboratories, New Mexico
P.O. Box 5800
Albuquerque, NM 87185-1413
Biography
Anne Ruffing joined Sandia in early 2010. As a Truman Fellow, Anne led her own research project on the Genetic Engineering of Cyanobacteria for Biodiesel Feedstock Production. In February of 2013, Anne converted to a technical staff position, in which she continues to research cyanobacteria and algae for biofuel production. Recent research in Anne’s laboratory applies metabolic engineering and synthetic biology approaches to develop novel biotechnologies for national security applications. Prior and ongoing projects in her laboratory include: (1) improving energy security by enabling photoautotrophic biofuel production, (2) developing novel biomonitoring technologies for the detection of chemical targets, and (3) applying synthetic biology for the development of biodefense technologies.
Education
Bachelor’s Degree: Chemical Engineering, University of Dayton (2000-2004)
Doctoral Degree: Chemical Engineering, Georgia Institute of Technology (2004-2010)
Postdoctoral Fellowships: President Harry S. Truman Fellowship in National Security Science and Engineering, Sandia National Laboratories (2010-2013)
During her graduate studies at the Georgia Institute of Technology, Ruffing worked with a polysaccharide-producing soil bacterium, Agrobacterium sp. ATCC 31749. Her thesis included metabolic engineering of the Agrobacterium for production of medically-important oligosaccharides, investigation of Agrobacterium physiology, and genomic and transcriptomic studies of polysaccharide production. As a Truman Fellow at Sandia National Laboratories, Ruffings’ work focused on the genetic modification of several cyanobacteria for the production of free fatty acids (FFAs) as feedstock for biodiesel production. This work characterized the physiological impact of FFA production, identified transcriptional changes associated with enhanced FFA production, and demonstrated the importance of cyanobacterial host selection.
Research Interests
Genetic Engineering of Cyanobacteria and Algae for Biofuel Production
Unlike eukaryotic algae, cyanobacteria are not natural lipid producers, yet cyanobacteria can be easily manipulated through genetic modification, have smaller genomes, and are regulated by prokaryotic genetics, making cyanobacteria ideal hosts for rational strain development. Ruffing’s research group has engineered the metabolisms of two model cyanobacteria, Synechococcus elongatus PCC 7942 and Synechococcus sp. PCC 7002, for the production of free fatty acids, a precursor for biodiesel production. The engineered strains were characterized using Sandia’s hyperspectral fluorescence imaging technology in addition to other traditional techniques, such as RNA-seq, to assess the effect of free fatty acid production on the cyanobacterium’s physiology and metabolism. The group seeks to continue to develop cyanobacteria for biofuel production through the application of high-throughput genome engineering technologies and integration with metabolic modeling and simulation.
Eukaryotic algae, such as Nannochloropsis gaditana, have shown promise for algal biofuel production, as they are natural lipid accumulators and demonstrate robust growth under outdoor production conditions. While additional strain development is necessary to achieve the desired characteristics for biofuel production, synthetic biology tools for genetic modification of eukaryotic algae are scarce. Ruffing’s group is currently developing tools for the genetic manipulation of N. gaditana, including acoustic-based delivery of DNA (i.e., sonoporation), characterized expression parts, and CRISPR-Cas9 editing technology. These tools will be used to engineer N. gaditana for robust growth under a range of environmentally relevant conditions.


Biosensors for the Detection of Chemicals
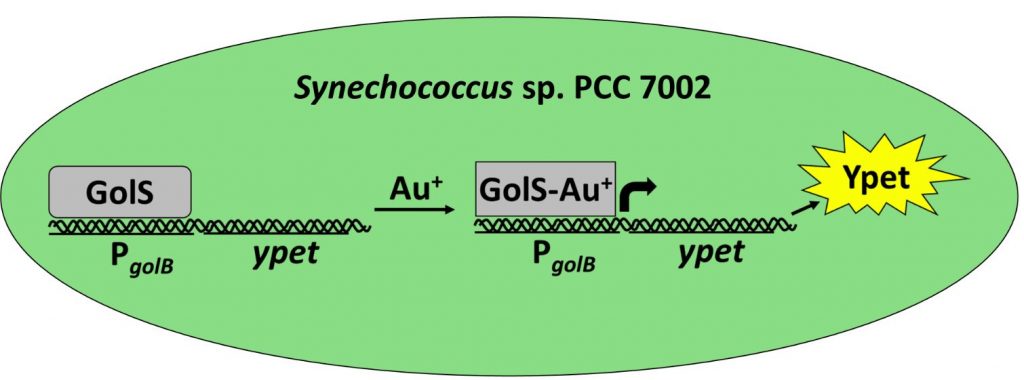
Biological organisms naturally sense chemicals in their environment through highly specific protein interactions. By leveraging and engineering these natural sensing systems, we can develop biological sensors that produce an easy-to-detect output signal in response to specific chemicals. Ruffing’s laboratory focuses on the development of biological sensors for the detection of metals, gases, and other small molecule targets. They use synthetic biology approaches to construct whole cell biosensors using natural proteins that have activity with a target chemical. This is combined with traditional and high-throughput protein evolution approaches to improve sensor function or evolve entirely novel detection capabilities. Additionally, they study natural responses in bacteria, algae, and plants to identify natural signatures that can be leveraged for chemical detection.
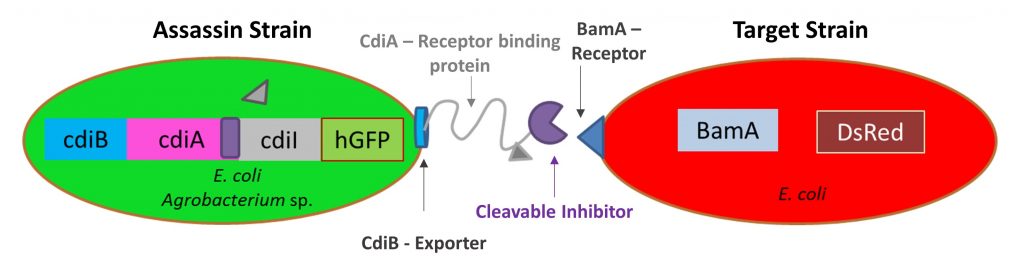
Engineering Natural Microbial Defense
Mechanisms to Inhibit Pathogenic Bacteria Bacteria have evolved natural mechanisms to inhibit or kill off competitor bacterial species that extend beyond traditional antibiotics. Ruffing’s group seeks to engineer bacterial systems with these natural inhibition mechanisms for selective targeting of pathogenic bacteria. Their work has primarily focused on engineering contact-dependent growth inhibition (CDI), a protein-based method of growth inhibition that has been demonstrated in several pathogenic bacteria. They aim to express heterologous CDI operons in non-pathogenic E. coli to selectively target and inhibit pathogenic bacteria, including Pseudomonas aeruginosa, Burkholderia pseudomallei, and Yersinia pestis.
Awards, Honors, and Memberships
- President Harry S. Truman Fellow in National Security Science and Engineering (2009)
- Philanthropic Educational Organization (P.E.O.) Scholar Award (2008-2009)
- Outstanding Teaching Assistant Award, Georgia Institute of Technology (2007)
- National Science Foundation Graduate Research Fellowship (2005-2008)
Publications
Raga Krishnakumar, Anne Ruffing, (2022). OperonSEQer: A set of machine-learning algorithms with threshold voting for detection of operon pairs using short-read RNA-sequencing data PLoS Computational Biology https://doi.org/10.1371/journal.pcbi.1009731 Publication ID: 79977
Anne Ruffing, Lucas Strickland, Patricia Gharagozloo, (2021). Sustainable Resources Inc. (NMSBA Closeout Report) https://doi.org/10.2172/1823240 Publication ID: 42185
Anne Ruffing, Joshua Podlevsky, Raga Krishnakumar, Chuck Smallwood, Tessa Dallo, Xavier Torres, Stephanie Kolker, John Morgan, Nathaphon King, Melissa Marsing, (2021). CERES: CRISPR Engineering for the Rapid Enhancement of Strains https://doi.org/10.2172/1820694 Publication ID: 75713
Danae Maes, August Finke, Chuck Smallwood, Jerilyn Timlin, Anne Ruffing, Michael Howard, (2021). Hyperspectral Bioindicators of Heavy Metal Exposure in Tall Fescue https://doi.org/10.2172/1884053 Publication ID: 79424
Anne Ruffing, Stephen Anthony, Lucas Strickland, Ian Lubkin, Carter Dietz, (2021). Identification of Metal Stresses in Arabidopsis thaliana Using Hyperspectral Reflectance Imaging Frontiers in Plant Science https://doi.org/10.3389/fpls.2021.624656 Publication ID: 76686
Tessily Hogancamp, Anne Ruffing, John Gladden, (2020). Developing RNA Interference in Rhodotorula toruloides for Gene Knockdown https://doi.org/10.2172/1837621 Publication ID: 72363
Tessily Hogancamp, Anne Ruffing, John Gladden, (2020). Developing RNA Interference in Rhodotorula toruloides for Gene Knockdown https://doi.org/10.2172/1837801 Publication ID: 72382
Anne Ruffing, Jesse Cahill, (2020). Exploring the Effect of Noble Gases on Enzyme Activity and Bacterial Growth https://doi.org/10.2172/1831599 Publication ID: 71586
Todd Lane, Anne Ruffing, John Gladden, Ramdane Harouaka, (2020). UT Austin Synthetic Biology Technical Exhange Presentation https://www.osti.gov/servlets/purl/1830932 Publication ID: 71394
Jesse Cahill, Anne Ruffing, (2020). Revisiting the Effects of Xenon on Urate Oxidase and Tissue Plasminogen Activator: No Evidence for Inhibition by Noble Gases Frontiers in Molecular Biosciences https://doi.org/10.3389/fmolb.2020.574477 Publication ID: 74743
Anne Ruffing, (2020). Challenges in Developing a Syn-Bio Toolkit for Microchloropsis gaditana https://www.osti.gov/servlets/purl/1804630 Publication ID: 73886
Chuck Smallwood, Anne Ruffing, Joshua Podlevsky, Raga Krishnakumar, Stephanie Kolker, Valerie Hines, (2019). Accelerating Photosynthetic Adaptations to Environmental Responses in Synechococcus sp. PCC 7002 https://www.osti.gov/servlets/purl/1642053 Publication ID: 64987
Anne Ruffing, Raga Krishnakumar, Chuck Smallwood, Joshua Podlevsky, Stephanie Kolker, Valerie Hines, (2019). Development of Genetic and Computational Tools for CRISPRi/a Screening in Synechococcus sp. PCC 7002 https://www.osti.gov/servlets/purl/1640704 Publication ID: 68882
Kara Peterson, Mauro Perego, Jennifer Frederick, Lauren Wheeler, Peter Bosler, Anne Ruffing, Ryan Davis, Leanne Whitmore, Kunal Poorey, Kelly Williams, Erika Roesler, Paul Bryan, Andrew Salinger, Jasper Hardesty, Vincent Tidwell, Anup Singh, (2018). SNL/BER Review 2018 https://www.osti.gov/servlets/purl/1594318 Publication ID: 59367
Chad Haddal, Diana Bull, Patricia Hernandez, Elizabeth Kistin-Keller, Brandon Heimer, Anne Ruffing, Munaf Aamir, (2018). Global Secuirty Implications of Chemical and Biological Innovation https://www.osti.gov/servlets/purl/1523711 Publication ID: 62285
Chad Haddal, Diana Bull, Patricia Hernandez, Elizabeth Kistin-Keller, Brandon Heimer, Anne Ruffing, Munaf Aamir, (2018). Global Security Implications of Chemical and Biological Innovation https://www.osti.gov/servlets/purl/1511995 Publication ID: 61932
Anne Ruffing, Dongmei Ye, Thomas Reichardt, Randy Lacey, (2018). PHOTOSYNTHETIC BIOSENSORS: ENGINEERING SYNECHOCOCCUS SP. PCC 7002 FOR THE DETECTION OF HEAVY METALS https://www.osti.gov/servlets/purl/1512382 Publication ID: 58576
Randy Lacey, Dongmei Ye, Anne Ruffing, (2017). Engineering Protein Based Whole Cell Biosensors for the Detection of Toxic Metals in the Environment https://www.osti.gov/servlets/purl/1474231 Publication ID: 53131
Anne Ruffing, Jordan McEwen, Lucas Strickland, Jaclyn Murton, Darren Branch, Eric Carnes, (2017). Genetic Tool Development for Nannochloropsis species https://www.osti.gov/servlets/purl/1648666 Publication ID: 56694
Randy Lacey, Dongmei Ye, Anne Ruffing, (2017). Protein Engineering for the Detection of Toxic Metals in the Environment https://www.osti.gov/servlets/purl/1458186 Publication ID: 56705
Anne Ruffing, Jordan McEwen, Todd Lane, Pamela Lane, Lucas Strickland, (2017). Engineering Green Algae for High Biomass Productivity https://www.osti.gov/servlets/purl/1418109 Publication ID: 52896
Anne Ruffing, Travis Jensen, Lucas Strickland, (2016). Genetic tools for advancement of Synechococcus sp. PCC 7002 as a cyanobacterial chassis Microbial Cell Factories https://doi.org/10.1186/s12934-016-0584-6 Publication ID: 52716
Anne Ruffing, George Bachand, Jerilyn Timlin, Ronald Manginell, Corey Hudson, Kelly Williams, Susan Rempe, C. Brinker, Maryla Olszewska-Wasiolek, Don Hanson, Victoria VanderNoot, (2016). SNL Capabilities for IV&V – DARPA BTO https://www.osti.gov/servlets/purl/1428154 Publication ID: 47973
Jordan McEwen, Anne Ruffing, Todd Lane, Pamela Lane, (2016). Genetic Tool Development for the green algae Nannochloropsis https://www.osti.gov/servlets/purl/1405255 Publication ID: 47310
Anne Ruffing, Todd Lane, Jordan McEwen, Pamela Lane, Lucas Strickland, (2016). Engineering Green Algae: Reducing Metabolic Waste for High Biomass Productivity https://www.osti.gov/servlets/purl/1375576 Publication ID: 51627
Anne Ruffing, Lucas Strickland, Travis Jensen, Jordan McEwen, Todd Lane, Pamela Lane, (2016). Genetic Tools for Engineering Cyanobacteria and Algae for Biofuel Production https://www.osti.gov/servlets/purl/1648722 Publication ID: 50924
Anne Ruffing, (2016). Anne Ruffing – Bio https://www.osti.gov/servlets/purl/1368642 Publication ID: 50597
Anne Ruffing, (2016). Metabolic Engineering and Systems Biology for Free Fatty Acid Production in Cyanobacteria https://www.osti.gov/biblio/1323888 Publication ID: 50232
Anne Ruffing, Todd Lane, Oliver Kilian, (2016). BETO Workshop Slides https://www.osti.gov/servlets/purl/1420859 Publication ID: 50060
Anne Ruffing, Travis Jensen, Lucas Strickland, (2016). Ionizing Radiation Resistance in Cyanobacteria https://www.osti.gov/servlets/purl/1367105 Publication ID: 49893
Anne Ruffing, Dongmei Ye, (2016). Engineering Two Component Signal Transduction in a Cyanobacterium https://www.osti.gov/servlets/purl/1339213 Publication ID: 46655
Anne Ruffing, Lucas Strickland, Travis Jensen, (2015). What makes cyanobacteria tick? Understanding promoter regulation in Synechococcus sp. PCC 7002 https://www.osti.gov/servlets/purl/1262939 Publication ID: 44348
Anne Ruffing, (2015). Cyanobacterial Biofuels: The Blue-Green Revolution https://www.osti.gov/servlets/purl/1249299 Publication ID: 43169
Anne Ruffing, Lucas Strickland, Travis Jensen, (2015). What makes cyanobacteria tick: Understanding promoter regulation in Synechococcus sp. PCC 7002 https://www.osti.gov/servlets/purl/1244440 Publication ID: 42583
Anne Ruffing, Travis jensen, Lucas Strickland, (2015). NMSBA: Aken Technologies Final Report: Toxicity Testing of Liquidoff https://doi.org/10.2172/1171600 Publication ID: 42186
Anne Ruffing, (2014). Towards the Development of a Cyanobacterial Chassis https://www.osti.gov/servlets/purl/1146946 Publication ID: 41138
Anne Ruffing, (2014). Biofuel Toxicity and Mechanisms of Biofuel Tolerance in Three Model Cyanobacteria Algal Research https://doi.org/10.1016/j.algal.2014.07.006 Publication ID: 40335
Anne Ruffing, (2014). Synechococcus sp. PCC 7002: A Cyanobacterial Chassis for Advanced Biofuel Production https://www.osti.gov/servlets/purl/1684925 Publication ID: 37287
Anne Ruffing, (2014). Improved Free Fatty Acid Production in Cyanobacteria with Synechococcus sp. PCC 7002 as Host Frontiers in Bioengineering and Biotechnology https://doi.org/10.3389/fbioe.2014.00017 Publication ID: 37872
Anne Ruffing, (2014). Systems-Level Synthetic Biology for Cyanobacterial Biofuel Production https://www.osti.gov/servlets/purl/1682596 Publication ID: 36810
Anne Ruffing, (2013). Eni Award 2014 Candidature Proposal https://www.osti.gov/servlets/purl/1673722 Publication ID: 36667
Anne Ruffing, (2013). Building a Synthetic Biology Toolbox for Synechococcus sp. PCC 7002 https://www.osti.gov/servlets/purl/1295564 Publication ID: 35696
Anne Ruffing, (2013). Pond scum to petrol : https://www.osti.gov/servlets/purl/1299210 Publication ID: 35073
Jerilyn Timlin, Aaron Collins, Anne Ruffing, (2013). Chemical Imaging of Cyanobacteria: A Picture is Worth a Thousand Words https://www.osti.gov/servlets/purl/1664738 Publication ID: 34974
Anne Ruffing, (2013). Synechococcus sp. PCC 7002 : https://www.osti.gov/servlets/purl/1506202 Publication ID: 34907
Anne Ruffing, (2013). Blue-Green Biofuels: Engineering Cyanobacterial Free Fatty Acid Production https://www.osti.gov/servlets/purl/1660823 Publication ID: 33067
Anne Ruffing, (2013). RNA-seq data for Gene Expression Omnibus https://www.osti.gov/servlets/purl/1658133 Publication ID: 32713
Anne Ruffing, (2013). Seq-ing Targets for Improving Free Fatty Acid Production in an Engineered Cyanobacterium Proposed for publication in Genome Biology. https://www.osti.gov/biblio/1072644 Publication ID: 32767
Anne Ruffing, Howland Jones, (2013). Genetic engineering of cyanobacteria as biodiesel feedstock https://doi.org/10.2172/1088046 Publication ID: 31504
Anne Ruffing, (2012). Metabolic Engineering of Cyanobacteria for Free Fatty Acid Production https://www.osti.gov/servlets/purl/1649894 Publication ID: 31110
Anne Ruffing, (2012). Borrowing Genes from Chlamydomonas reinhardtii for Free Fatty Acid Production in Engineered Cyanobacteria Proposed for publication in Metabolic Engineering. https://www.osti.gov/biblio/1062257 Publication ID: 30586
Jerilyn Timlin, Brian Dwyer, Christine Trahan, Omar Garcia, Kylea Parchert, Anne Ruffing, Howland Jones, Thomas Reichardt, Amy Powell, Aaron Collins, Patricia Gharagozloo, (2012). From benchtop to raceway : spectroscopic signatures of dynamic biological processes in algal communities https://doi.org/10.2172/1055623 Publication ID: 30300
Christine Trahan, Anne Ruffing, (2012). Biofuels Toxicity in Freshwater and Marine Model Cyanobacterial Strains https://www.osti.gov/servlets/purl/1647998 Publication ID: 30186
Thomas Reichardt, Aaron Collins, Anne Ruffing, Howland Jones, Brian Dwyer, Jerilyn Timlin, (2012). Remote spectroradiometric monitoring of Nannochloropsis salina in a fluidically mixed pond https://www.osti.gov/biblio/1067567 Publication ID: 28497
Anne Ruffing, (2012). Borrowing Genes from Green Algae to Improve Cyanobacterial Free Fatty Acid Production https://www.osti.gov/biblio/1064200 Publication ID: 28547
Anne Ruffing, Christine Trahan, Howland Jones, (2012). Blue-Green Biofuels: Engineering Cyanobacteria for Fuel Production https://www.osti.gov/servlets/purl/1650271 Publication ID: 28879
Jerilyn Timlin, Kylea Parchert, Lindsey Hughes, Aaron Collins, Thomas Reichardt, Howland Jones, Amy Powell, Brian Dwyer, Anne Ruffing, (2012). Optimizing Algal Cultivation: An Innovative Multidiscipline and Multiscale Approach https://www.osti.gov/servlets/purl/1141076 Publication ID: 28628
Anne Ruffing, (2012). Fuel-producing cyanobacteria cry foul https://www.osti.gov/servlets/purl/1650478 Publication ID: 28235
Anne Ruffing, Howland Jones, (2012). Physiological Effects of Free Fatty Acid Production in Genetically Engineered Synechococcus elongatus PCC 7942 Proposed for publication in Biotechnology and Bioengineering. https://doi.org/10.1002/bit.24509 Publication ID: 26211
Anne Ruffing, (2012). Engineering Cyanobacteria for Biofuel Production: New Host New Challenges https://www.osti.gov/servlets/purl/1648463 Publication ID: 25864
Anne Ruffing, (2011). Expression of novel genes from Chlamydomonas reinhardtii in a cyanobacterium for enhanced production of free fatty acids https://www.osti.gov/servlets/purl/1288667 Publication ID: 25000
Anne Ruffing, (2011). Engineering Green Fuel: Genetic Engineering of Cyanobacteria for the Production of Biodiesel Feedstocks https://www.osti.gov/servlets/purl/1663322 Publication ID: 24403
Anne Ruffing, Michelle Aragon, Omar Garcia, Howland Jones, (2011). Genetic Engineering of Cyanobacteria as Biodiesel Feedstock https://www.osti.gov/servlets/purl/1662112 Publication ID: 24096
Anne Ruffing, Michelle Aragon, Omar Garcia, Howland Jones, (2011). Characterization of genetically engineered Synechococcus elongatus PCC 7942 for biofuel production https://www.osti.gov/servlets/purl/1288484 Publication ID: 23490
Anne Ruffing, Aaron Collins, Omar Garcia, Kylea Parchert, Howland Jones, Jerilyn Timlin, Amy Powell, (2011). Programmed cell death-like responses in Chlamydomonas reinhardtii https://www.osti.gov/servlets/purl/1289697 Publication ID: 23491
Thomas Reichardt, Omar Garcia, Aaron Collins, Anne Ruffing, Howland Jones, Michelle Aragon, Brian Dwyer, Christine Trahan, Jerilyn Timlin, (2011). Remote spectraradiometric monitoring of Nannochloropsis salina growth https://www.osti.gov/servlets/purl/1289796 Publication ID: 23536
Anne Ruffing, Michelle Aragon, Omar Garcia, Howland Jones, (2011). Genetic Engineering and Characterization of Cyanobacteria for Biodiesel Production https://www.osti.gov/biblio/1108247 Publication ID: 22606
Aaron Collins, Thomas Reichardt, Anne Ruffing, Christine Trahan, Brian Dwyer, Omar Garcia, Lindsey Hughes, Howland Jones, Todd Lane, Laura Carney, Amy Powell, (2011). Pond-Scale Diagnostics in Algae Production https://www.osti.gov/servlets/purl/1109317 Publication ID: 22908
Anne Ruffing, (2011). Engineered Cyanobacteria: Teaching an Old Bug New Tricks Bioengineered Bugs https://doi.org/10.4161/bbug.2.3.15285 Publication ID: 21255
Anne Ruffing, Michelle Aragon, Howland Jones, (2010). Metabolic engineering of synechococcus elongatus PCC 7942 for free fatty acid production https://www.osti.gov/biblio/1030278 Publication ID: 20618
Anne Ruffing, (2010). Genetic engineering of cyanobacteria for biodiesel feedstock https://www.osti.gov/biblio/1028329 Publication ID: 20224
Jerilyn Timlin, Brian Dwyer, Howland Jones, Aaron Collins, Anne Ruffing, Thomas Reichardt, Omar Garcia, James Ricken, Amy Powell, Christine Trahan, (2010). Optimizing Algal Cultivation & Productivity: An Innovative Multidiscipline and Multiscale Approach https://www.osti.gov/servlets/purl/1732236 Publication ID: 20110
Jerilyn Timlin, Omar Garcia, Michelle Aragon, Amy Powell, Howland Jones, Thomas Reichardt, James Ricken, Christine Trahan, Anne Ruffing, Aaron Collins, Brian Dwyer, (2010). From benchtop to raceway : spectroscopic signatures of dynamic biological processes in algal communities https://www.osti.gov/biblio/1028414 Publication ID: 19578
Anne Ruffing, (2010). Metabolic Engineering of Cyanobacteria as Biodiesel Feedstock https://www.osti.gov/servlets/purl/1678882 Publication ID: 18554
Jerilyn Timlin, Howland Jones, James Ricken, Jaclyn Murton, Brian Dwyer, Anne Ruffing, Amy Powell, Thomas Reichardt, (2010). Optimizing algal cultivation & productivity : an innovative, multidiscipline, and multiscale approach https://www.osti.gov/biblio/1000993 Publication ID: 17893
Showing Results.
Selected Publications
- Haddal CC, Bull DL, Pacheco Hernandez PM, Kistin Keller EJ, Heimer BW, Ruffing AM, and Aamir MS. Global security implications of chemical and biological innovation. European Commission Joint Research Centre’s Future Technology Assessment Conference Proceedings. June 2018. Brussels, Belgium.
- Ruffing AM, Jensen TJ, and Strickland LM. Genetic tools for advancement of Synechococcus sp. PCC 7002 as a cyanobacterial chassis. Microbial Cell Factories. 2016. 15:190. doi: 10.1186/s12934-016-0584-6
- Ruffing AM and Kallas T. Editorial: Cyanobacteria: The Green E. coli. Frontiers in Bioengineering and Biotechnology. 2016. Research Topic ebook. 4:7. doi: 10.3389/fbioe.2016.00007
- Ruffing AM and Trahan CA. Biofuel toxicity and mechanisms of biofuel tolerance in three model cyanobacteria. Algal Research. 2014. 5:121-132.
- Ruffing AM. Improved free fatty acid production in cyanobacteria with Synechococcus sp. PCC 7002 as host. Frontiers in Bioengineering & Biotechnology. 2014. 2:17.
- Ruffing AM. RNA-Seq analysis and targeted mutagenesis for improved free fatty acid production in an engineered cyanobacterium. Biotechnology for Biofuels. 2013. 6:113.
- Ruffing AM. Borrowing genes from Chlamydomonas reinhardtii for free fatty acid production in engineered cyanobacteria. Journal of Applied Phycology. 2013. 25(5): 1495-1507.
- Ruffing AM and Jones HDT. Physiological effects of free fatty acid production in genetically engineered Synechococcus elongatus PCC 7942. Biotechnology and Bioengineering. 2012. 109(9): 2190-2199. (Cover, Spotlight)
- Ruffing AM and Chen RR. Transcriptome profiling of a curdlan-producing Agrobacterium reveals conserved regulatory mechanisms of exopolysaccharide biosynthesis. Microbial Cell Factories. 2012. 11:17. Reichardt TA, Garcia OF, Collins AM, Jones HDT,
- Ruffing AM, and Timlin JA. Spectroradiometric monitoring of Nannochloropsis salina growth. Algal Research. 2012. 1(1): 22-31.
- Ruffing AM. Engineered Cyanobacteria: Teaching an old bug new tricks. Bioengineered Bugs. 2011. 2(3).
- Ruffing AM, Castro-Melchor M, Hu W-S, and Chen RR. Genome sequencing of the curdlan-producing Agrobacterium sp. ATCC 31749. Journal of Bacteriology. 2011. 193(16): 4294-4295.
- Ruffing AM and Chen RR. Citrate stimulates oligosaccharide synthesis in metabolically engineered Agrobacterium sp. Applied Biochemistry and Biotechnology. 2011. 164(6): 851-866.
- Ruffing AM and Chen RR. Metabolic engineering of Agrobacterium sp. strain ATCC 31749 for production of an α-Gal epitope. Microbial Cell Factories. 2010. 9(1).
- Ruffing AM and Chen RR. Metabolic engineering of microbes for oligosaccharide and polysaccharide synthesis. Microbial Cell Factories. 2006. 5: 25-33.
- Ruffing AM, Mao Z, and Chen RR. Metabolic engineering of Agrobacterium sp. for UDP-galactose regeneration and oligosaccharide synthesis. Metabolic Engineering. 2006. 8(5): 465-473.
Selected Book Chapters
- Ruffing AM. Metabolic Engineering and Systems Biology for Free Fatty Acid Production in Cyanobacteria. Book chapter in Cyanobacteria: Omics and Manipulation. Editor: Dmitry A. Los. Caister Academic Press. Poole, UK. 2017. p161-186. doi: 10.21775/9781910190555.08.
- Ruffing AM. Metabolic Engineering of Hydrocarbon Biosynthesis for Biofuel Production. Book chapter in Liquid, Gaseous and Solid Biofuels – Conversion Techniques. Intech, Rijeka, Croatia. 2013.
- Ruffing AM and Chen RR. Metabolic Engineering of Microorganisms for Oligosaccharide and Polysaccharide Production. Book chapter in Microbial Production of Biopolymers and Polymer Precursors: Applications and Perspectives. Horizon Bioscience, Wymondham, UK. 2009. p197-228.
- Ruffing AM and Chen RR. Metabolic Engineering and Other Methods of Strain Improvement. Book chapter in Advances in Fermentation Technology. Asiatech Publishers, Inc. New Delhi. 2008. p119-144.
Selected Patents
- Reichardt TA, Collins AM, Garcia OF, Ruffing AM, Timlin JA, and Jones HDT. Spectroradiometric Monitoring of Algal Growth. 2013. US patent application: 13869854.
- Carnes E, Ruffing AM, and Ashley C. Delivery Platforms for the Domestication of Algae and Plants. 2015. US patent application: 14/871,748.